 » NMR Jobs
» NMR Jobs |
|
|
|
|
|
|
 » Cool papers
» Cool papers |
|
|
|
|
|
|
 » NMR books
» NMR books |
|
|
|
|
|
|
 » NMR theses
» NMR theses |
|
|
|
|
|
|
 » NMR blogs
» NMR blogs |
|
|
|
|
|
|
 » NMR news
» NMR news |
|
|
|
|
|
|
 » NMR tweets
» NMR tweets |
|
|
|
|
|
|
 » NMR videos
» NMR videos |
|
|
|
|
|
|
 » NMR pictures
» NMR pictures |
|
|
|
|
|
|
 » Online Users: 306
» Online Users: 306 |
| 0 members and 306 guests |
| No Members online |
| Most users ever online was 1,278, 01-09-2024 at 07:38 AM. |
|
 » Welcome!
» Welcome! |
Welcome, NMR world!
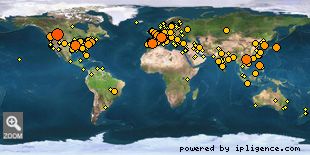
Our visitors map.
|

 Sparse pseudocontact shift NMR data obtained from a non-canonical amino acid-linked lanthanide tag improves integral membrane protein structure prediction Sparse pseudocontact shift NMR data obtained from a non-canonical amino acid-linked lanthanide tag improves integral membrane protein structure prediction |
|
Apr 05, 2023 - 5:15 PM - by nmrlearner
|
 Sparse pseudocontact shift NMR data obtained from a non-canonical amino acid-linked lanthanide tag improves integral membrane protein structure prediction
Sparse pseudocontact shift NMR data obtained from a non-canonical amino acid-linked lanthanide tag improves integral membrane protein structure prediction
Abstract
A single experimental method alone often fails to provide the resolution, accuracy, and coverage needed to model integral membrane proteins (IMPs). Integrating computation with experimental data is a powerful approach to supplement missing structural information with atomic detail. We combine RosettaNMR with experimentally-derived paramagnetic NMR restraints to guide membrane protein structure prediction. We demonstrate this approach using the disulfide bond formation protein B (DsbB), an α-helical IMP. Here, we attached a cyclen-based paramagnetic lanthanide tag to an engineered non-canonical amino acid (ncAA) using a copper-catalyzed azide-alkyne cycloaddition (CuAAC) click chemistry reaction. Using this tagging strategy, we collected 203 backbone HN pseudocontact shifts (PCSs) for three different labeling sites and used these as input to guide de novo membrane protein structure prediction protocols in Rosetta. We find that this sparse PCS dataset combined with 44 long-range NOEs as restraints in our calculations improves structure prediction of DsbB by enhancements in model accuracy, sampling, and scoring. The inclusion of this PCS dataset improved the Cα-RMSD transmembrane segment values of the best-scoring and best-RMSD models from 9.57Â*Ã? and 3.06Â*Ã? (no NMR data) to 5.73Â*Ã? and 2.18Â*Ã?, respectively.
... [Read More]
|

 0 Replies | 293 Views
0 Replies | 293 Views
|
|
 » BioNMR wiki
» BioNMR wiki |
|
|
|
|
|
|
 » NMR discussion
» NMR discussion |
|
|
|
|
|
|
 » NMR conferences
» NMR conferences |
|
|
|
|
|
|
 » NMR software
» NMR software |
|
|
|
|
|
|
 » Pulse sequences
» Pulse sequences |
|
|
|
|
|
|
 » NMR community
» NMR community |
|
|
|
|
|
|
 » NMR presentations
» NMR presentations |
|
|
 » NMR web resources
» NMR web resources |
|
|
|
|
|
|
 » NMR feature requests
» NMR feature requests |
|
|
 » NMR bookmarks
» NMR bookmarks |
|
|
|
|
|
|
 » Stats
» Stats |
Members: 3,202
Threads: 25,741
Posts: 26,127
Top Poster: nmrlearner (23,185)
|
| Welcome to our newest member, bpadmanabhan |
|



