Watergate vs Presaturation
Biochemists and protein chemists are often interested in observing the NH protons in their samples. Since the NH protons usually undergo slow chemical exchange with water, it is desirable to run the samples in H2O rather than D2O so the NH protons will not exchange with the deuterium in the solvent which would make them
invisible in the 1H NMR spectrum. In practice, a mixture of 10% D2O and 90% H2O is used as a solvent so that a deuterium lock can be established and used while running the spectrum. The very high concentration of water compared to the very low concentration of solute necessitates the use of solvent suppression methods.
Both
presaturation and
WATERGATE are efficient techniques used to suppress strong water signals from proton NMR spectra, however, there are differences between the two methods of which the user must be aware. Presaturation employs a selective, long, low power pulse to saturate the water resonance. This pulse is usually several seconds in duration during which exchange can occur between the unsaturated NH protons and the saturated water protons. If this exchange occurs, the NH protons become partially saturated to an extent related to the rate of chemical exchange between the NH and the water. The intensity of the NH protons in the spectrum is non-quantitative. WATERGATE, on the other hand, uses a pair of gradients surrounding a composite pulse which in effect inverts all but the water signal. The duration of the composite pulse is about 4 orders of magnitude shorter than a presaturation pulse, so exchange between the NH protons and the solvent occurs to a much lesser extent during the WATERGATE sequence compared to presaturation. As a result, the NH region is much less attenuated and more quantitative in a spectrum collected using WATERGATE compared to a similar spectrum run with presaturation. This is illustrated in the figure below which shows the NH region of the 1H spectrum of a small disaccharide substituted peptide.

One can see that some of the NH's are greatly attenuated in the spectrum acquired with presaturation compared to the spectrum run using WATERGATE. The more attenuated the signal, the faster the chemical exchange for that particular NH with water.
The WATERGATE suppression sequence is very similar to the gradient spin echo sequences used to measure diffusion constants and
DOSY spectra. As a result, when WATERGATE suppression is used, one expects diffusion losses for small molecules (which diffuse quickly) compared to large molecules (which diffuse slowly). This effect is demonstrated in the figure below which shows the NH/aromatic region of the 1H spectrum of a partially degraded 15N labelled 10 kDa protein.
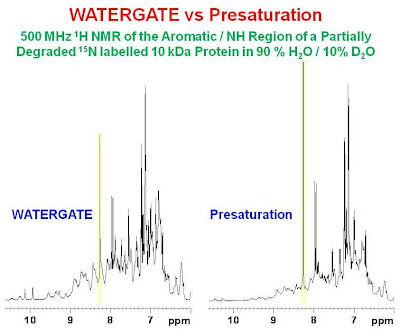
Here, one can see the same exchange losses pointed out in the previous figure for the presaturation spectrum compared to the WATERGATE spectrum. In addition, one can see that the intensity of a very sharp peak marked in yellow (likely due to a CH proton from free histidine) is less intense in the WATERGATE spectrum compared to the presaturation spectrum. This loss is due to the fast diffusion of the small free amino acid compared to the very large protein. In conclusion, one must be aware of the differences between the two solvent suppression methods if quantitative results are being sought.
I would like to thank Roger Tam from
Robert Ben's Laboratory and Allison Sherratt from
Natalie Goto's Laboratory for kindly providing the samples of the peptide and protein, respectively.



Source:
University of Ottawa NMR Facility Blog



