Learning NMR with Deep Purple
It’s not that I’ve gone crazy (well, I hope not :-) ) or that Deep Purple has moved from Hard Rock to Science (or at least I‘m not aware of this), but after
my last post about the acoustic reproduction of NMR FIDS, I thought that it would be fun to compose a simple song and, at the same time, create some stuff which can serve as an educational tool for some very basic NMR.
I won’t actually compose anything original, but rather make an NMR cover of the famous Smoke on the Water riff by Deep Purple. It’s very simple with a central theme consisting in a four-note "blues scale" melody. If you don’t know this song, you can watch and listen here:
These are the notes for the central theme I learnt by ear. I know it’s not 100% accurate, but for the purpose of the exercise this should do just fine:
DFG DFAG DFG FD
You can play this riff with the virtual piano below, just click on the picture and click the notes above. It’s fun!
 NMRing Smoke on the water
NMRing Smoke on the water
Before any further ado, these are the instructions to listen to the NMR version of Smoke on the Water:
- First you will need Mnova. If you don’t own a license, you can download a free, fully functional demo from our web site.
- Download this NMR document (SmokeOnTheWater.mnova)
- Download this script (playArray.qs)
- Finally, put on your earphones or amplify the volume on your computer speakers, open SmokeOnTheWater.mnova file with Mnova and run the script.
If everything works as I hope, you should be listening to 12 different pings pretending to be the melody of Smoke on the Water riff. I hope you won’t get too disappointed and more importantly, I hope that Deep Purple won’t detest me for ruining their song :)
Behind the scenes
As Dr. Walter Bauer explained on his web site, the most convenient way to create melodies with NMR is by means of pulse programmers to define the appropriate frequencies and delays. I’m taking a much simpler approach which, of course, cannot be used to create complex harmonies.
The basic idea is to synthesize NMR FIDs in such a way that each FID will represent a note of the song. For example, in this particular case I have simulated 4 FIDS with frequencies corresponding to A, D,F, and G. Next I stacked these 4 FIDs to create a new stacked item with 12 FIDs formed by the combination of the 4 main FIDs (main tones) and sorted to yield DFG DFAG DFG FD. The result is illustrated in the figure below.
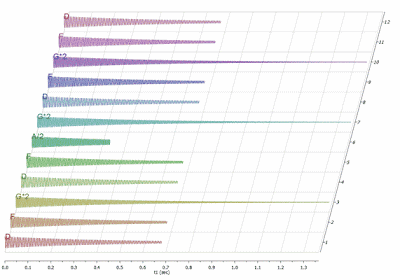
As you can see, not all the FIDs have the same length. This is, of course, because some notes have to last longer than others. For example, we can consider the first FID (D) as the quarter note (crotchet), the third FID a half note (minim) and the sixth FID an eighth note (quaver). Next, I will comment on how the duration of the FIDs are controlled.
Some points of interest
At first sight, it may seem that in order to create the NMR version of Smoke on the Water one simply has to create the 4 FIDs with the ‘real’ frequencies corresponding to A, D, F and G notes and then organize them accordingly. For example, using the A220 pitch, the frequencies for the different notes should be:
A = 220 Hz
D = 146.83
F = 174.61
G = 196 Hz
However, if these notes are used in this way, the resulting song will be totally different than expected! The reason for that is very simple: NMR FIDs are expected to be in the so-called Quadrature Detection mode, that is, zero frequency in the center of the spectral width. Thus, it becomes necessary to translate the original frequencies into the NMR quadrature frame. The equation for such transformation is very simple:
NMR Note Frequency = Note Frequency + SpectralWidth/2
For example, in this case, as I have used a spectral width of 3000 Hz, the NMR frequencies for the 4 notes will be:
A = 220 + 3000/2 = 1720 Hz
D = 146.83 + 3000/2 = 1646.83 Hz
F = 174.61 + 3000/2 = 1674.61 Hz
G = 196 + 3000/2 = 1696 Hz
Another point worth mentioning concerns the way to modulate the duration of the different FIDs to create crotchets, quavers, etc. In NMR terms, this is equivalent to the acquisition time (AT) which is defined as:
AT = N / SW
Where N is the number of points and SW is the spectral width. We can modify any of these two values to change the duration of the note (= FID acquisition time). In this example, I have kept the spectral width constant and modified the number of points.
Finally …
In this example I have created pure tone notes (e.g. one single frequency for each note) but it could also be possible to create some kind of guitar chords (e.g. power chords) to make the song more realistic by simply combining the root tone and a fifth. This can be easily done by using the Spin Simulation toolkit available in Mnova.
If you haven’t had enough of Deep Purple yet, you can watch a Jazz version of Smoke in the Water in the video below. Enjoy!
Please note that if you are using Mnova under Windows 7 or Linux, you may not be able to play the song as described earlier on. I am currently investigating the reason for this, as tests carried out on other Windows and Mac versions went smoothly. Should you experience these difficulties,
you can listen to the song as a .WAV file here.
 More...
More...
Source:
NMR-analysis blog


