Installing the DOSY toolbox on a Mac
The DOSY toolbox is an open source program written by Dr. Mathias Nilsson that runs on Windows, Linux, Mac and on any other platform covered by MATLAB. I am going to review it next week, while today I am giving a few practical tips that I prefer to leave out of the review, for the sake of readability and tidiness.
I know that 14% of my readers have a Mac, and I have verified that the original installation instructions of the toolbox are inaccurate and unclear (for the average Mac user at least), so today I will simply explain how you can install the DOSY toolbox on your Mac.
Download the file called DOSYToolbox06_MAC_13May09_pkg from the address:
http://personalpages.manchester.ac.u...n/software.htm
With a double click this archive generates a folder called: "DOSYToolbox06_MAC_13May09_pkg 2". This name is actually too long and useless, therefore I have shortened it into "DOSYToolbox06" and moved the folder inside my home directory. This is not strictly necessary but is a safe and sensible move. Inside the folder there is an installer called "MCRInstaller.dmg". It installs the runtime of MATLAB, which is necessary (unless you already own MATLAB itself). The difference is... technically speaking... to make it short... well the difference is $2,000 that you save if you install the runtime instead of buying matlab itself. Got it? A little slower but much cheaper.
The installer works like any other installer: you must always say "yes", you can't be wrong, eventually you will read it's all OK but you have no clue of what it has done and this is terribly bad because you absolutely need to know where the files have been installed, otherwise you can't run the DOSY toolbox at all. Fortunately I have discovered where the files have gone:
/Applications/MATLAB/MATLAB_Compiler_Runtime/v710
(this is valid, of course, for version 7.1 of the runtime).
The downloaded folder also contains a script called: "run_DOSYToolbox06_MAC.sh". This is the command that starts the program, and it needs an argument and this argument is the path to the matlab runtime that you have already installed. In other words, the canonical way to start the toolbox would be to type a command like this into the Terminal:
/path/to/run_DOSYToolbox06_MAC.sh /path/to/MATLAB_Compiler_Runtime/v710
for example:
/Users/your_name/DOSYToolbox06/run_DOSYToolbox06_MAC.sh /Applications/MATLAB/MATLAB_Compiler_Runtime/v710
This is highly impractical, but you can easily create a double-clickable shortcut:
- open the Automator application
- choose the template called "workflow"
- into the leftmost column, click on "Utilities"
- into the second column, double click on "Run Shell Script"
- from the pop-up menu "Shell" choose "/bin/bash"
- fill the main field with our command:
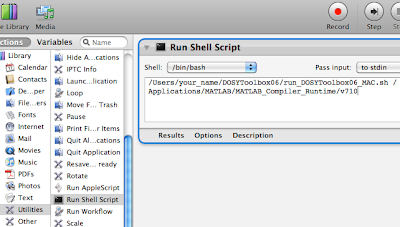
Run this workflow. If, in a few seconds, the DOSY toolbox window appears, it means that the workflow works. Quit the DOSY toolbox and save the workflow. Be careful and select the proper File Format: it should be "Application" and not "Workflow".
Finally you have created a double-clickable application (you can paste a different icon)

and you can forget Terminal, shell, bash and the likes...
At the time of this writing, though, it seems that the DOSY toolbox is fully functional only if launched from a shell. It is a puzzling and unwelcome incident, I hope it's going to find a solution soon.
Has anybody the compiler for the Mac? Would this person be so kind to compile version 0.7 for the rest of us Mac users? "Open source" means that some times you should also give, not always take.
 More...
More...



